rDNA Variability and Group I Introns
As late as the 1990s it had been assumed that all SSU rDNA genes were highly conserved with little sequence variation between species of closely related genera. Yet I found that using different PCR primer pairs on DNA isolated from a fungal culture produced PCR fragments from the same region differing in size by up to 450 nt. These extra nucleotides resulted from discrete insertions in the SSU rDNA. P. T. DePriest discovered similar insertions in the SSU rDNA isolated from species of the pixie cup lichen Cladonia sp., and determined that these were group I introns. Upon comparing our data we discovered many identical insertion locations. On exploring further we found that some insertion locations appear to be ancient and conserved between organisms as distantly related as fungi, green algae, and amoebae although there may be little sequence homology between these introns' DNA sequences. Other introns may be of more recent origin. In our analyses of over one hundred SSU rDNA sequences from various ascolichens we found variation in intron occurrence within species, within populations, and possibly within rDNA repeats in a single tandem array.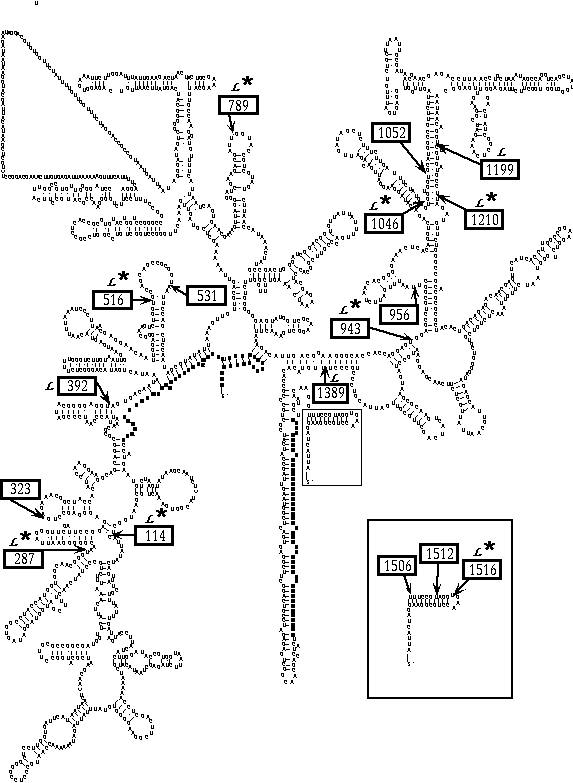 Insertions, most of which have been identified as group I introns, occur in at least 17 positions in highly conserved regions of the SSU rDNA in fungi, algae, and plastids. Introns of similar size were located at identical locations within the SSU rDNA genes we had sequenced. Paula T. DePriest and I published a list of all currently reported positions of insertions and group I introns in the SSU rDNA Molecular Biology and Evolution 12: 208-218; download PDF).
Lichen-forming fungi have a surprisingly large number of insertions in their SSU rDNA; a culture of Lecanora dispersa contains eight insertions within its SSU rDNA, doubling the typical size of 1,800 nt. DePriest and I found variation within species, within populations, and possibly, within rDNA repeats in a single tandem array. For a closer view of this secondary structure please see this descriptive page.
Insertions, most of which have been identified as group I introns, occur in at least 17 positions in highly conserved regions of the SSU rDNA in fungi, algae, and plastids. Introns of similar size were located at identical locations within the SSU rDNA genes we had sequenced. Paula T. DePriest and I published a list of all currently reported positions of insertions and group I introns in the SSU rDNA Molecular Biology and Evolution 12: 208-218; download PDF).
Lichen-forming fungi have a surprisingly large number of insertions in their SSU rDNA; a culture of Lecanora dispersa contains eight insertions within its SSU rDNA, doubling the typical size of 1,800 nt. DePriest and I found variation within species, within populations, and possibly, within rDNA repeats in a single tandem array. For a closer view of this secondary structure please see this descriptive page.
ITS2 rDNA Secondary Structure
Another assumption was that the Internal Transcribed Spacer regions (ITS1 & ITS2) of ribosomal DNA were simply junk DNA. Although these sequences had been useful identifying markers, they could not be aligned between fungal samples at taxonomic levels much higher than genera. I found that the ITS2 region has a conserved secondary structure, reminiscent of that found in group I introns. Fungal ITS2 sequences can usually be folded into four loops, with those the two loops closest to the 5.8S more conserved. Using these loop structure as guides it is possible to align ITS2 rDNA sequences among Eumycota. We developed methods using these loop regions for taxonomy and systematics.Landis FC and A Gargas. 2007. Using ITS2 secondary structure to create species-specific oligonucleotide probes for fungi.
Mycologia 99: 681-692. ![]() download preprint PDF (180 kb)
download preprint PDF (180 kb)